ChloroScan: A metagenomic workflow to recover chloroplast genomes.
Author: Yuhao Tong (The University of Melbourne)
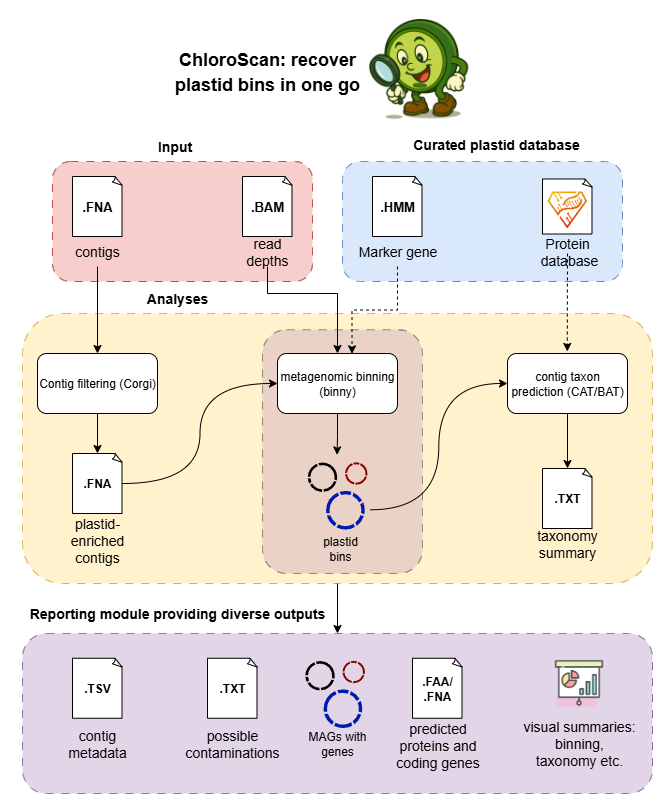
A snakemake workflow for MMA metagenomics for recovering chloroplast genomes.
This workflow is designed to recover chloroplast genomes from metagenomic data.
In order to run ChloroScan properly, we recommend you to setup conda environment in your data directory otherwise it may exceed your home directory’s quota if not much space is left.
The space size to setup ChloroScan (conda environments and source codes) is about 14G. While after installing configuration database the minimum space required may be ~100G.
Installation
To install the workflow, use pip3 (recommended). Firstly create a virtual environment.
python3 -m venv chloroscan_env
source chloroscan_env/bin/activate
pip3 install chloroscan
The newest update is v0.1.5. Currently we don’t plan to deploy it on conda.
Configuration database
Besides installation, ChloroScan requires several databases: one for taxonomy classification by CAT and another for inferring plastid MAG qualities by binning.
1. CAT database
Your choices can be either using the CAT-prepared database which contains all proteins in NCBI’s nr database. However, it is ram-taking and time-consuming when running CAT, and requires ~300GB of disk.
To assess plastid contig taxonomy, we recommend downloading our curated protein database consisting of all plastid genome-encoded proteins from NCBI and proteins from UniRef90 (size: ~85GB after unzipping).
command to download:
wget --referer=https://figshare.unimelb.edu.au --user-agent="Mozilla/5.0" -O "CAT_db.tar.gz" https://figshare.unimelb.edu.au/ndownloader/files/51053993
2. Binny database
You don’t need to download it manually, ChloroScan will load it once you installed the conda environment for each job.